Discover how a bimodal integration strategy can address the major data management challenges facing your organization today.
Get the Report →Analyze Greenhouse Data in R
Use standard R functions and the development environment of your choice to analyze Greenhouse data with the CData JDBC Driver for Greenhouse.
Access Greenhouse data with pure R script and standard SQL on any machine where R and Java can be installed. You can use the CData JDBC Driver for Greenhouse and the RJDBC package to work with remote Greenhouse data in R. By using the CData Driver, you are leveraging a driver written for industry-proven standards to access your data in the popular, open-source R language. This article shows how to use the driver to execute SQL queries to Greenhouse and visualize Greenhouse data by calling standard R functions.
Install R
You can match the driver's performance gains from multi-threading and managed code by running the multithreaded Microsoft R Open or by running open R linked with the BLAS/LAPACK libraries. This article uses Microsoft R Open 3.2.3, which is preconfigured to install packages from the Jan. 1, 2016 snapshot of the CRAN repository. This snapshot ensures reproducibility.
Load the RJDBC Package
To use the driver, download the RJDBC package. After installing the RJDBC package, the following line loads the package:
library(RJDBC)
Connect to Greenhouse as a JDBC Data Source
You will need the following information to connect to Greenhouse as a JDBC data source:
- Driver Class: Set this to cdata.jdbc.greenhouse.GreenhouseDriver
- Classpath: Set this to the location of the driver JAR. By default this is the lib subfolder of the installation folder.
The DBI functions, such as dbConnect and dbSendQuery, provide a unified interface for writing data access code in R. Use the following line to initialize a DBI driver that can make JDBC requests to the CData JDBC Driver for Greenhouse:
driver <- JDBC(driverClass = "cdata.jdbc.greenhouse.GreenhouseDriver", classPath = "MyInstallationDir\lib\cdata.jdbc.greenhouse.jar", identifier.quote = "'")
You can now use DBI functions to connect to Greenhouse and execute SQL queries. Initialize the JDBC connection with the dbConnect function.
You need an API key to connect to Greenhouse. To create an API key, follow the steps below:
- Click the Configure icon in the navigation bar and locate Dev Center on the left.
- Select API Credential Management.
- Click Create New API Key.
- Set "API Type" to Harvest.
- Set "Partner" to custom.
- Optionally, provide a description.
- Proceed to Manage permissions and select the appropriate permissions based on the resources you want to access through the driver.
- Copy the created key and set APIKey to that value.
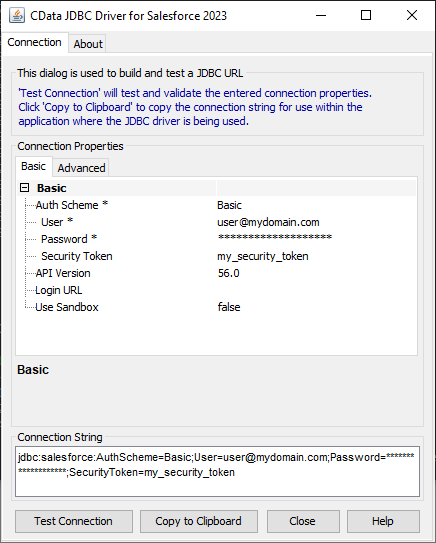
Built-in Connection String Designer
For assistance in constructing the JDBC URL, use the connection string designer built into the Greenhouse JDBC Driver. Either double-click the JAR file or execute the jar file from the command-line.
java -jar cdata.jdbc.greenhouse.jar
Fill in the connection properties and copy the connection string to the clipboard.
Below is a sample dbConnect call, including a typical JDBC connection string:
conn <- dbConnect(driver,"jdbc:greenhouse:APIKey=YourAPIKey;")
Schema Discovery
The driver models Greenhouse APIs as relational tables, views, and stored procedures. Use the following line to retrieve the list of tables:
dbListTables(conn)
Execute SQL Queries
You can use the dbGetQuery function to execute any SQL query supported by the Greenhouse API:
applications <- dbGetQuery(conn,"SELECT Id, CandidateId FROM Applications WHERE Status = 'Active'")
You can view the results in a data viewer window with the following command:
View(applications)
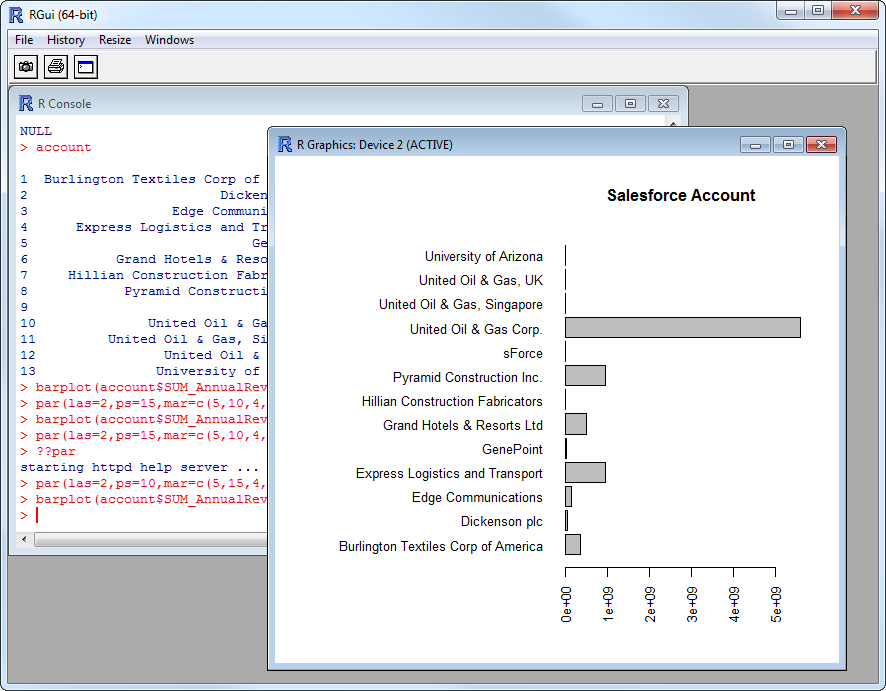
Plot Greenhouse Data
You can now analyze Greenhouse data with any of the data visualization packages available in the CRAN repository. You can create simple bar plots with the built-in bar plot function:
par(las=2,ps=10,mar=c(5,15,4,2))
barplot(applications$CandidateId, main="Greenhouse Applications", names.arg = applications$Id, horiz=TRUE)